3.1 Sleep deprivation exacerbates circadian and behavioral disturbances in SiNPs-treated mice
After two weeks of therapy, pallet and water consumption, physique weight, core physique temperature, and locomotor exercise had been assessed within the NC, SiNPs-treated, and SD + SiNPs-treated teams. Locomotor exercise, monitored through telemetry, revealed notable alterations within the SiNPs-treated and SD + SiNPs-treated mice in comparison with controls (Figs. 2A-B). In the course of the daytime (ZT0-ZT12), the SD + SiNPs-treated group exhibited larger ranges of exercise than the NC and SiNPs-treated teams. Conversely, throughout the nighttime (ZT12-ZT24), the SD + SiNPs-treated group’s locomotor exercise decreased to decrease ranges, whereas the NC and SiNPs-treated teams confirmed elevated exercise ranges that had been larger than these of the SD + SiNPs group. Though the SiNPs-treated group adopted an identical 24-hour pattern to the NC group, its common exercise stage over the day was decrease than that of the NC group. The every day rhythms of the SiNPs-treated and SD + SiNPs-treated teams additionally differed considerably from the NC group (Figs. 2A-B). The core physique temperature of the NC group adopted a standard circadian sample, growing with locomotor exercise throughout the darkish cycle (Fig. 2C). In distinction, the SiNPs-treated and SD + SiNPs-treated group confirmed an irregular sample, their core physique temperature was decrease than the NC group in each the sunshine and darkish cycles (Figs. 2C-D).
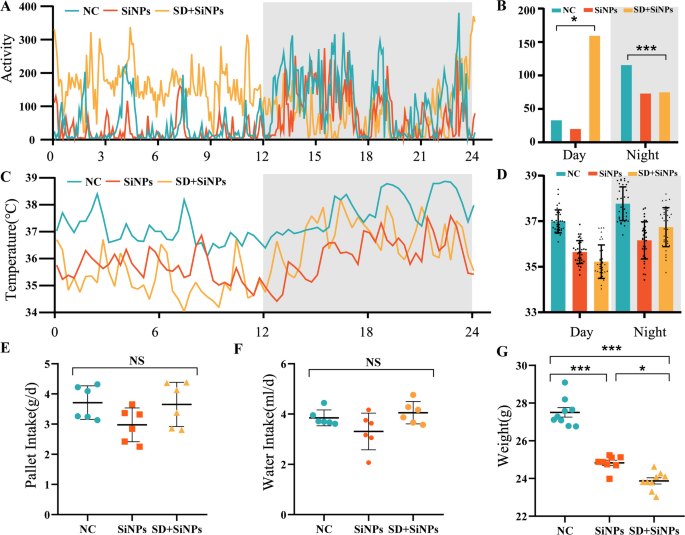
Behavioral and physiological alterations in mice following SiNPs and SD + SiNPs remedies. (A-B) Locomotor exercise patterns over a 24-hour interval within the NC, SiNPs-treated, and SD + SiNPs-treated teams. Information had been collected each 5 min. The grey shading signifies the darkish part. *P < 0.05, ***P < 0.001. (C-D) Core physique temperature fluctuations over a 24-hour interval within the NC, SiNPs-treated, and SD + SiNPs-treated teams. Information had been recorded each 20 min. The grey shading signifies the darkish part. *P < 0.05, ***P < 0.001. (E) Pellet consumption within the NC, SiNPs-treated, and SD + SiNPs-treated teams. NS: not important. (F) Water consumption within the NC, SiNPs-treated, and SD + SiNPs-treated teams. NS: not important. (G) Physique weight modifications within the NC, SiNPs-treated, and SD + SiNPs-treated teams. *P < 0.05, ***P < 0.001
To evaluate the impact of SD on the final behaviors of the mice, we collected knowledge on pellet consumption, water consumption, and physique weight over a two-week interval. We noticed no important statistical variations in water consumption and pellet consumption among the many three teams (Fig. 2E-F). Pellet and water consumption had been barely decrease within the SD + SiNPs-treated group in comparison with the NC group, however the distinction was not statistically important. The SiNPs-treated group exhibited the bottom consumption among the many three teams, but these reductions had been additionally not statistically important (Fig. 2E-F). Regardless of the dearth of great variations in water consumption, there was a transparent pattern in each pellet and water consumption, in addition to a big distinction in physique weight, with the SD + SiNPs-treated group weighing lower than the opposite two teams (Fig. 2G).
Collectively, these outcomes display that SD considerably exacerbates SiNPs-induced disturbances in circadian habits and physiology, as evidenced by alterations in locomotor exercise, core temperature regulation, and physique weight.
3.2 Structural alterations within the ELGs triggered by sleep deprivation in SiNPs-treated mice
Our earlier research have proven that the scale and weight of ELGs exhibit circadian oscillations, which may be modulated by each intrinsic (e.g., circadian disruption) [33] and extrinsic elements (e.g., sort 1 diabetes) [48]. Constructing upon these findings, we investigated whether or not publicity to SiNPs and SD, individually or together, may alter the physiological traits of ELGs, significantly their construction and performance.
ELG weights had been measured at 4 ZT factors (ZT0, ZT6, ZT12, and ZT18) throughout all experimental teams. Within the NC group, ELG weight displayed strong diurnal variation, peaking at ZT18. This rhythmicity was abolished in each the SiNPs-treated and SD + SiNPs-treated teams. Notably, SD additional exacerbated the disruption attributable to SiNPs (Fig. 3A). Total, ELG weights had been considerably decrease in each therapy teams in comparison with NC, indicating compromised lacrimal gland homeostasis (Fig. 3B).
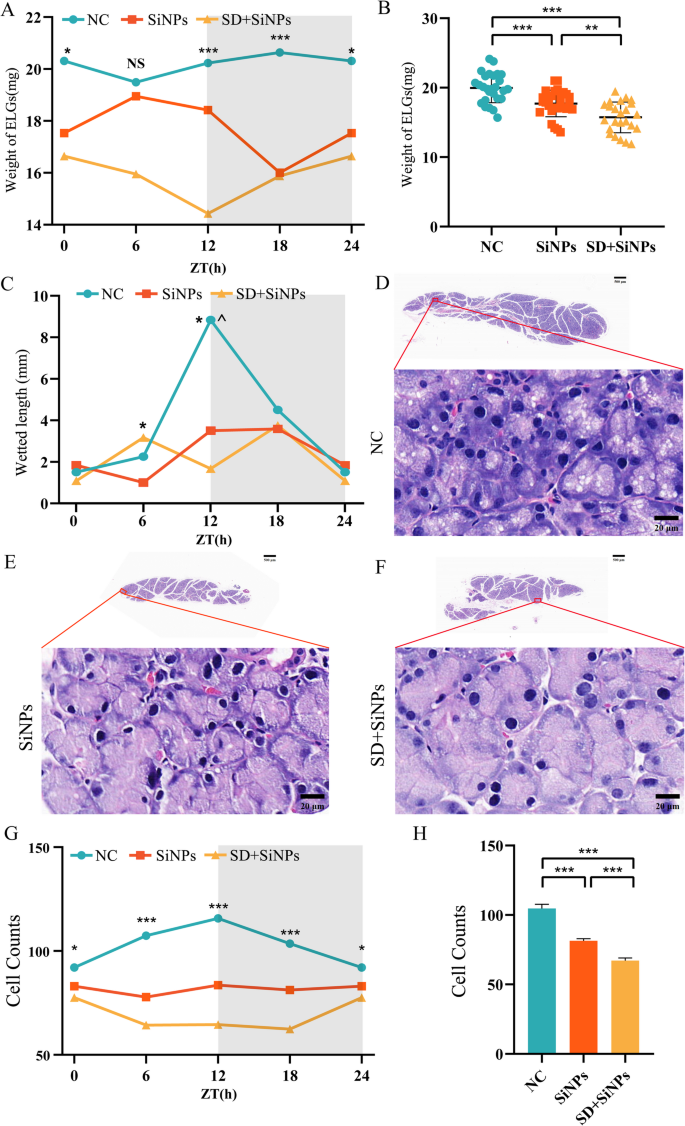
Influence of SiNPs and SD + SiNPs therapy on ELG weight, tear secretion, and structural integrity. (A) Diurnal modifications of ELG weight within the NC, SiNPs-treated, and SD + SiNPs-treated teams. *P < 0.05, ***P < 0.001. NS: not important. (B) ELG weight measurements within the NC, SiNPs-treated, and SD + SiNPs-treated teams. **P < 0.01, ***P < 0.001. (C) Tear secretion was assessed utilizing the phenol thread take a look at at ZT 0, 6, 12, and 18 over a 24-hour cycle. Statistical significance is denoted as *P < 0.05 for comparisons between SiNPs-treated and SD + SiNPs-treated teams and ^P < 0.05 for comparisons between the NC and SD + SiNPs-treated teams. (D-F) Consultant gross sections of ELGs from the NC (D), SiNPs-treated (E), and SD + SiNPs-treated (F) teams (scale bar: 500 μm, higher panels). Magnified views of the boxed areas present the acinar cell morphology (scale bar: 20 μm, decrease panels). (G) Diurnal modifications of ELG cell counts within the NC, SiNPs-treated, and SD + SiNPs-treated teams. *P < 0.05, ***P < 0.001. (H) Quantitative evaluation of acinar cell counts within the NC, SiNPs-treated, and SD + SiNPs-treated teams. Statistical evaluation was carried out utilizing Brown-Forsythe ANOVA. A number of comparisons had been performed utilizing the Video games-Howell submit hoc take a look at. ***P < 0.001
Tear secretion, was assessed utilizing phenol crimson thread assessments, additionally adopted a circadian rhythm within the NC group, with a peak at ZT18. This rhythmic sample was absent within the SiNPs-treated and SD + SiNPs-treated teams. Importantly, no statistically important variations in tear quantity had been noticed between the 2 therapy teams, suggesting that SiNPs-induced dysfunction had already reached a plateau, with SD (Fig. 3C).
To evaluate structural modifications within the lacrimal gland, HE staining was carried out for histopathological examination (Figs. 3D-F). Determine 3G exhibits every day oscillations within the NC group, which is disrupted in each the SiNPs-treated and SD + SiNPs-treated teams. Quantitative evaluation in Fig. 3H revealed important variations within the variety of cell nuclei among the many three teams. The variety of nuclei was considerably decreased within the SiNPs-treated group in comparison with the NC group, and additional decreased within the SD + SiNPs-treated group relative to each the NC group and the SiNPs-treated group. These findings recommend that each SiNPs publicity and the mixed intervention with SD induce structural abnormalities and purposeful impairment in ELGs, with extra extreme results noticed within the SD + SiNPs-treated group.
Collectively, these findings display that SD intensifies SiNPs-evoked disruptions in locomotor exercise, core temperature regulation, and physique weight.
3.3 Alterations in international gene expression of ELGs promoted by sleep deprivation in SiNPs-treated mice
To research the oscillations within the transcriptome of murine ELGs, we collected ELGs at three-hour intervals all through a 24-hour interval from the NC, SiNPs-treated, and SD + SiNPs-treated teams of C57BL/6J mice. RNA-seq was carried out on the BGISEQ-500 platform, and transcriptome evaluation was performed utilizing the JTK_CYCLE algorithm with a 24-hour oscillation interval and a significance threshold of P < 0.05.
A complete of 20,242 genes had been recognized by means of RNA-seq evaluation. Genes had been categorized into rhythmic (FPKM ≥ 0.1, adjusted P < 0.05), non-rhythmic (FPKM ≥ 0.1, adjusted P ≥ 0.05), and low-expressed (FPKM < 0.1) based mostly on the obtained knowledge. Among the many 20,242 transcripts, rhythmic genes constituted 15.10%, 13.50%, and 16.26% of the NC, SiNPs-treated, and SD + SiNPs-treated teams, respectively. Low-expressed genes accounted for 33.30%, 33.02%, and 34.22% within the respective teams. Non-rhythmic genes had been current in 51.60%, 53.48%, and 49.52% of the NC, SiNPs-treated, and SD + SiNPs-treated teams (Figs. 4A-C).
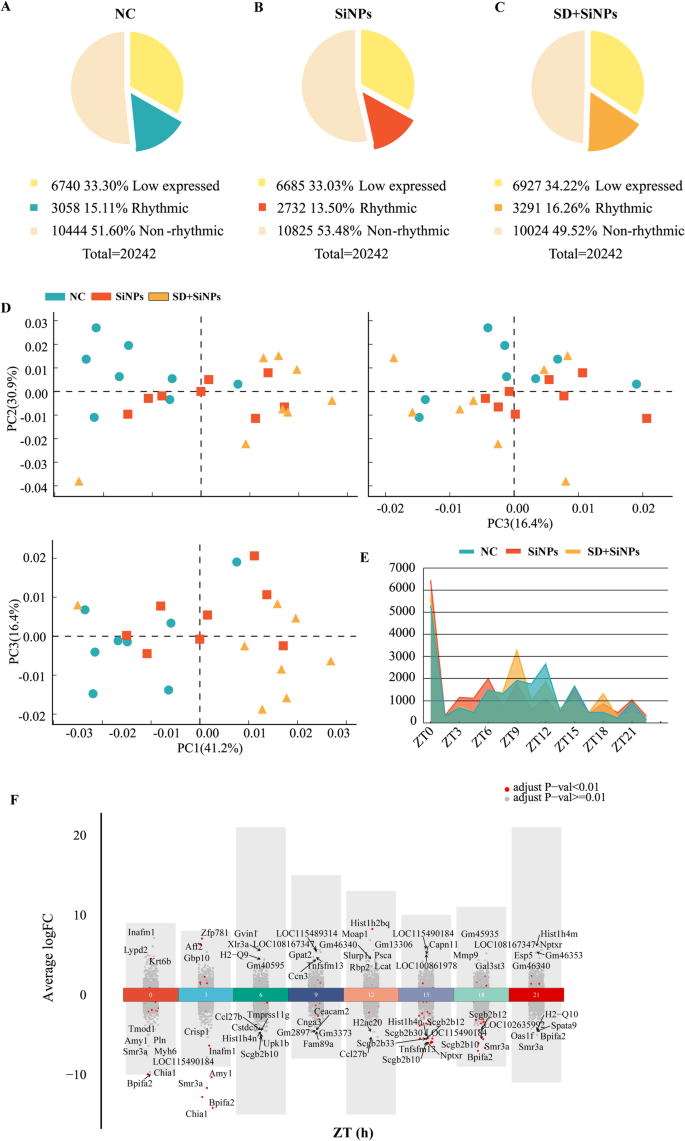
Comparative transcriptomic evaluation of mouse ELGs in response to SiNPs and SD + SiNPs remedies. (A–C) The pie charts illustrate the transcriptomic composition of ELGs within the NC, SiNPs-treated, and SD + SiNPs-treated teams, categorized into low-expressed, rhythmic, and non-rhythmic genes. (D) The PCA scatterplot depicts the general gene expression profiles of ELGs within the NC (blue), SiNPs-treated (crimson), and SD + SiNPs-treated (yellow) teams. Every dot represents a person animal sampled at three-hour intervals over a 24-hour cycle (N = 3 per time level). The shaded areas point out the distribution of every group. (E) The road graph illustrates the temporal distribution of peak gene expression throughout totally different ZT factors within the NC, SiNPs-treated, and SD + SiNPs-treated teams. Variations in peak expression instances spotlight the influence of remedies on rhythmic gene regulation. (F) The volcano plot visualizes DEGs in ELGs between NC and SiNPs-treated mice. The x-axis represents ZT factors, whereas the y-axis signifies fold change (FC). Purple and grey dots denote genes with adjusted P-values < 0.01 and ≥ 0.01, respectively. Every group consisted of 24 mice
In comparison with the NC group, the SiNPs-treated group skilled a lower of 0.27% in low-expressed genes and 1.61% in rhythmic genes, whereas the SD + SiNPs-treated group confirmed a rise of 0.92% in low-expressed genes and 1.16% in rhythmic genes. The proportion of non-rhythmic genes elevated by 1.88% within the SiNPs-treated group and decreased by 2.48% within the SD + SiNPs-treated group relative to NC (Figs. 4A-C).
Principal element evaluation (PCA) was employed to discern distinct groupings among the many remedies. The primary three principal parts accounted for 41.2%, 30.9%, and 16.4% of the variance, respectively (Fig. 4D). The PCA biplots demonstrated that the NC, SiNPs-treated, and SD + SiNPs-treated teams fashioned three distinct clusters, indicating that SiNPs and SD + SiNPs remedies have a big influence on differential gene expression.
The distribution of gene expression peaks over a 24-hour interval revealed the results of SiNPs and SD + SiNPs remedies (Fig. 4E). The NC group exhibited peak gene expression at ZT12 and ZT15. In distinction, the SiNPs-treated group displayed peaks at ZT0, ZT3, ZT6, and ZT21. The SD + SiNPs-treated group confirmed peak expression at ZT9 and ZT18.
Volcano plot successfully highlights the top10 considerably differentially expressed genes (DEGs) between the NC group and the SiNPs-treated group (Fig. 4F and Desk S1), offering insights into the impact of SiNPs therapy on ELGs. Volcano plots of great DEGs between the NC group and the SD + SiNPs-treated, and between the SiNPs-treated and the SD + SiNPs-treated, are introduced in Determine S2 and Tables S2-3.
Total, these findings display SiNPs publicity—accentuated by SD—considerably alters the ELG transcriptome, differentially affecting rhythmic and non-rhythmic genes and highlighting the circadian sensitivity of the lacrimal gland to environmental and behavioral stressors.
3.4 Disruption of circadian rhythmicity of ELGs promoted by sleep deprivation in SiNPs-treated mice
To discover the influence of SiNPs therapy and SD + SiNPs therapy on the circadian rhythmicity of ELGs, we employed Venn diagrams, heatmaps, pie charts, and rose diagrams to research the rhythmic genes among the many three teams. Venn diagrams had been utilized to determine distinctive and shared rhythmic genes throughout the NC, SiNPs-treated, and SD + SiNPs-treated teams. Among the many 5,901 non-redundant gene transcripts, the proportion of distinctive rhythmic genes in every group was 18.53% for the NC, 17.32% for the SiNPs-treated, and 23.88% for the SD + SiNPs-treated teams (Fig. 5A and Desk S4).
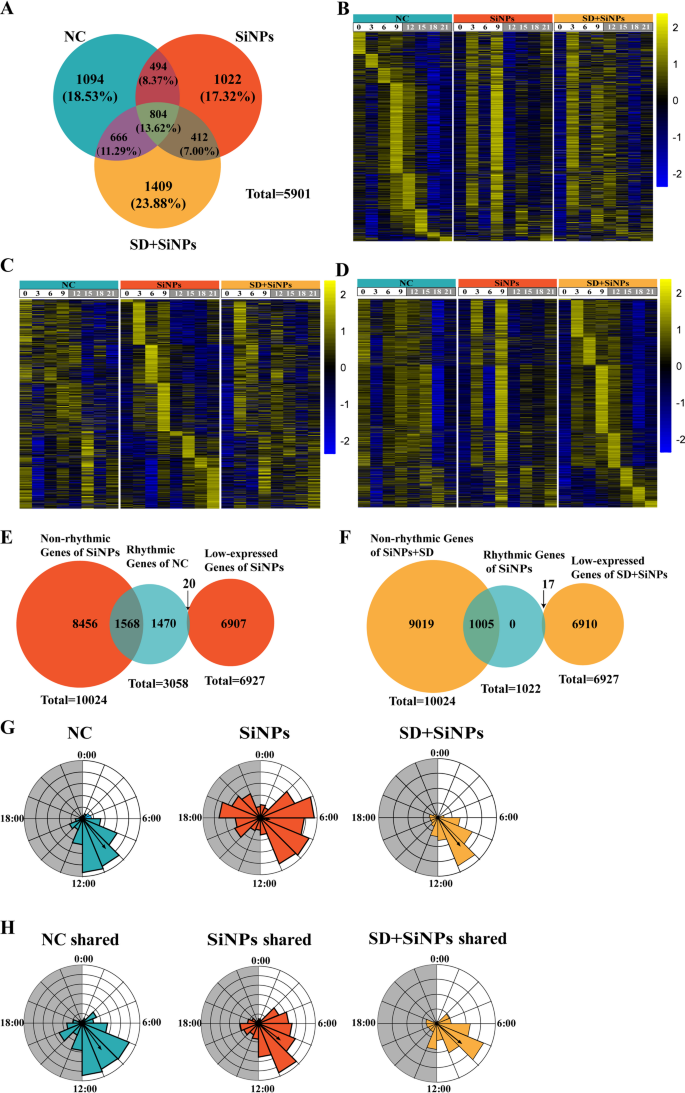
Results of SiNPs and SD + SiNPs therapy on the rhythmic transcriptome of murine ELGs. (A) The venn diagram illustrates the overlap and divergence of rhythmic transcripts among the many NC, SiNPs-treated, and SD + SiNPs-treated teams. (B) The heatmaps show the expression profiles of 1,094 rhythmic transcripts distinctive to the NC group at totally different ZT factors. The left panel represents the NC group, whereas the center and proper panels correspond to the SiNPs-treated and SD + SiNPs-treated teams, respectively. Gene expression ranges are normalized inside a ± 2 vary, as indicated by the colour scale. (C) The heatmaps present the expression ranges of 1,022 rhythmic transcripts unique to the SiNPs-treated group throughout numerous ZT factors. The panel association is per (B), with the NC group on the left, the SiNPs-treated group within the center, and the SD + SiNPs-treated group on the proper. (D) The heatmaps illustrates the expression patterns of 1,409 rhythmic transcripts distinctive to the SD + SiNPs-treated group over a number of ZT factors, following the identical panel association as in (B) and (C). (E) The venn diagram depicts the transition of rhythmic genes within the NC group to both non-rhythmic (blue) or low-expressed (crimson) genes within the SiNPs-treated group. (F) The venn diagram illustrates the conversion of rhythmic genes within the SiNPs-treated group into non-rhythmic (crimson) or low-expressed (yellow) genes within the SD + SiNPs-treated group. (G-H) The wind rose diagrams signify the imply vector and size of rhythmic genes distinctive to every group (G) and people shared throughout all three teams (H). The NC, SiNPs-treated, and SD + SiNPs-treated teams are positioned on the proper, center, and left, respectively
Heatmaps had been constructed for instance the expression ranges of 1,096 distinctive genes within the NC group (Figs. 5B), 1,022 within the SiNPs-treated group (Figs. 5C), and 1,409 within the SD + SiNPs-treated group (Figs. 5D) throughout eight ZT factors. The heatmaps revealed important alterations in gene expression patterns between the SiNPs-treated group and the NC group, with additional pronounced modifications noticed within the SD + SiNPs-treated group.
Pie charts had been used to depict the transformation of rhythmic genes post-treatment (Figs. 5E-F). Of the rhythmic genes distinctive to NC group, 1,470 maintained their rhythmic expression after SiNPs therapy, whereas 1,568 transitioned to non-rhythmic standing (Fig. 5E). Within the SiNPs-treated group, not one of the distinctive rhythmic genes remained unchanged after SD + SiNPs therapy; 1,005 grew to become non-rhythmic, and 17 grew to become low-expressed (Fig. 5F).
The rose diagrams, generated utilizing Oriana software program, visually demonstrated the variations within the periodicity and part distribution of distinctive and shared rhythmic genes throughout teams. The NC group exhibited a imply vector (µ) of 9:30 and a imply vector size (r) of 0.635, the SiNPs-treated group confirmed µ = 6:40 and r = 0.163, and the SD + SiNPs-treated group introduced µ = 9:26 and r = 0.596 (Fig. 5G). Determine 5H illustrates the modifications within the common vector and its size for shared rhythmic genes beneath totally different remedies: NC group (µ = 9:37, r = 0.55), SiNPs-treated group (µ = 8:29, r = 0.463), and SD + SiNPs-treated group (µ = 8:34, r = 0.532).
The composite pie charts revealed part shifts amongst shared genes throughout teams. After SiNPs therapy, 33.83% of shared genes within the NC group maintained their part, whereas 66.17% skilled part shifts, with 68.98% advancing and 31.02% delaying. Following SD + SiNPs therapy, 34.45% of shared genes remained phase-stable, 65.55% underwent part modifications, with 43.83% delaying and 56.17% advancing. Within the SD + SiNPs therapy group, 82.71% of shared genes exhibited part alterations, 66.32% superior, and 33.68% delayed, with solely 17.29% remaining unchanged (Fig. S1).
The findings point out that each SiNPs therapy and the mixture of SD + SiNPs therapy considerably disrupt the circadian rhythmicity of ELGs, resulting in substantial alterations within the expression and part distribution of rhythmic genes, which underscores the profound influence of those remedies on the molecular clock of lacrimal gland operate.
3.5 Reshaped KEGG and phase-set enriched pathways triggered by sleep deprivation in SiNPs-treated mice
To judge the purposeful implications of circadian gene expression modifications, KEGG pathway enrichment evaluation was carried out on rhythmic genes distinctive to the NC, SiNPs-treated, and SD + SiNPs-treated teams. The genes unique to the NC group had been considerably enriched in 9 pathways (Fig. 6A and Desk S5, P < 0.05), with a give attention to mobile processes and metabolic pathways. Nevertheless, the rhythmic genes of SiNPs-treated group enriched in several KEGG pathways from the NC group (Fig. 6B and Desk S6, P < 0.05), so did the rhythmic genes of SD + SiNPs-treated group (Fig. 6C and Desk S7, Q < 0.05). Notably, the cell cycle pathway was the one pathway shared between the SiNPs-treated and SD + SiNPs-treated teams.
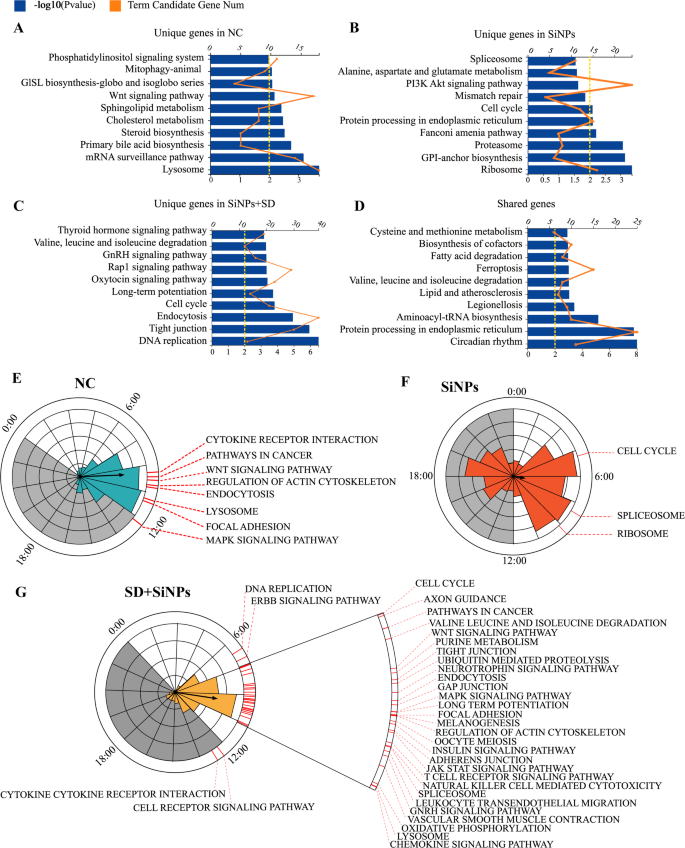
Influence of SiNPs and SD + SiNPs therapy on KEGG and phase-clustered pathways in mouse ELGs. (A–D) Gene annotation of KEGG pathways considerably enriched in rhythmic genes distinctive to the NC group (A), SiNPs-treated group (B), SD + SiNPs-treated group (C), and people shared amongst all three teams (D), with P < 0.01. The highest 10 enriched pathways are introduced. The higher horizontal axis, aligned with the orange line graph, represents the variety of time period candidate genes, whereas the decrease horizontal axis, aligned with the blue histogram, represents the -log10 (P-value), indicating the statistical significance of enrichment. (E–G) Abstract of considerably phase-clustered pathways (P < 0.05) distinctive to the NC group (E), SiNPs-treated group (F), and SD + SiNPs-treated group (G). The interior circle and column size signify the part distribution of rhythmic genes particular to every group. The outer crimson line marks KEGG pathways (P < 0.05) related to rhythmic genes distinctive to every group, indicating enrichment at distinct ZT factors, as decided by the part distribution within the interior circle. Grey shading signifies darkish cycles
A complete of 18 pathways had been enriched amongst genes widespread to all teams (Fig. 6D and Desk S8, Q < 0.05). The highest 10 pathways recognized for every group had been categorized into 5 teams: Mobile Processes, Genetic Info Processing, Metabolism, Organismal Methods, and Environmental Info Processing. The SiNPs-treated group’s pathways fell inside Mobile Processes, Genetic Info Processing, and Metabolism (Desk S5). After the SD + SiNPs therapy, the enriched pathways expanded to incorporate all 5 classes (Desk S6). Though the SD + SiNPs-treated group’s distinctive pathways matched the management group’s classes, the SD + SiNPs therapy didn’t totally counteract the results of SiNPs therapy (Desk S7). In line with KEGG pathway stage 2, the organic course of classes for the SD + SiNPs-treated group diverged from these of the NC and SiNPs-treated teams, suggesting that SD + SiNPs therapy could amplify the results of SiNPs therapy.
PSEA was employed to research the temporal expression patterns of those pathways. The NC group’s considerably enriched pathways had been predominantly expressed between ZT9 and ZT12 (Fig. 6E and Desk S9, Kuiper Q < 0.05). In distinction, the SiNPs-treated group had three pathways peaking close to ZT4, ZT5, and ZT9 (Fig. 6F and Desk S10, Kuiper Q < 0.05). The SD + SiNPs-treated group displayed enriched pathway expression primarily between ZT6 and ZT12, with two pathways, B cell receptor signaling and cytokine receptor interplay, exhibiting expression throughout the darkish part (Fig. 6G and Desk S11, Kuiper Q < 0.05).
Total, these findings display that SiNPs disturb the part distribution of a number of ELG pathways, and SD broadens and deepens these shifts, exhibiting that mixed environmental and behavioral stressors can significantly compromise circadian and purposeful homeostasis within the lacrimal gland.
3.6 Alterations in cluster-dependent transcriptomic map and KEGG pathways triggered by sleep deprivation in SiNPs-treated mice
To research the temporal tendencies in rhythmic gene expression, we utilized the Mfuzz gentle clustering bundle for evaluation. Our findings revealed that genes from the NC group (Figs. 7A-D), the SiNPs-treated group (Figs. 7E-H), the SD + SiNPs-treated group (Figs. 7I-L) had been every categorized into 4 distinct clusters, every exhibiting distinctive expression profiles and enriched pathways.
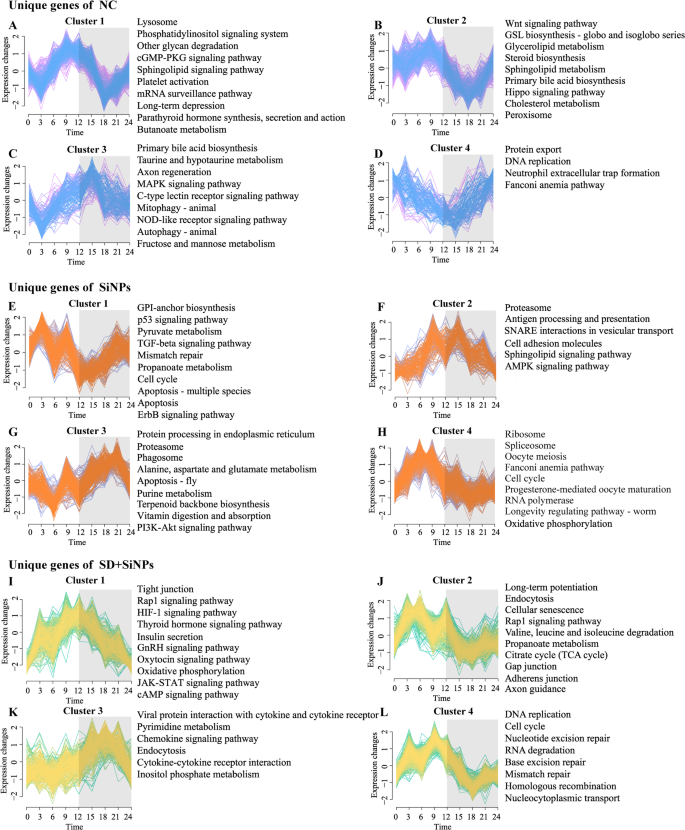
Influence of SiNPs and SD + SiNPs therapy on the circadian gene clustering profile and KEGG pathways in murine ELGs. (A–L) Temporal gene expression Z-scores for 4 distinct enriched clusters distinctive to the NC group (A–D), SiNPs-treated group (E–H), and SD + SiNPs-treated group (I–L). Blue, orange, and yellow strains signify genes with low membership values, whereas pink, purple, and inexperienced strains point out genes with excessive membership values. Grey shading denotes darkish cycles. The corresponding right-side panels show the highest 10 KEGG pathways (P < 0.05) enriched for circadian genes distinctive to every group and cluster
In Cluster 1, important disparities had been noticed among the many NC, SiNPs-treated, and SD + SiNPs-treated teams, with gene counts of 439, 242, and 289, respectively. The NC group exhibited a notably larger variety of clustered genes. The genes within the NC group peaked at ZT9 and troughed at ZT3 and ZT18 (Fig. 7A, left). In distinction, the SiNPs-treated group peaked at ZT3 and troughed at ZT12 (Fig. 7E, left), whereas the SD + SiNPs-treated group peaked at ZT12 (Fig. 7I, left). These distinct international patterns underscore the divergent regulatory mechanisms at play.
Cluster 2 confirmed 367, 157, and 503 genes within the NC, SiNPs-treated, and SD + SiNPs-treated teams, respectively. The NC and SD + SiNPs-treated teams shared related expression patterns, peaking at ZT9 and troughing at ZT18 (Figs. 7B and F, left). Nevertheless, the SiNPs-treated group deviated from this pattern, peaking at ZT12 and troughing at ZT3 (Fig. 7J, left).
The gene expression patterns in Cluster 3 had been largely concordant throughout the teams, with gene counts of 165, 269, and 231, respectively, albeit with variations within the timing of peaks and troughs (Figs. 7C, G and Okay, left). In Cluster 4, the SiNPs-treated and SD + SiNPs-treated teams exhibited related gene counts (358 and 392, respectively) (Figs. 7H and L, left), in distinction to the NC group, which had a considerably decrease rely of 127 (Fig. 7D, left). The expression patterns of enriched genes within the SiNPs-treated and SD + SiNPs-treated teams had been broadly related however diverged from these of the NC group.
Moreover, the KEGG pathways enriched in genes belonging to every cluster different throughout teams, indicating a reprogramming of the transcriptomic panorama in response to SiNPs therapy and SD (Figs. 7A-L, proper). The KEGG pathways related to the NC, SiNPs-treated, and SD + SiNPs-treated teams, together with their respective clusters, are proven in Tables S12-14, respectively. This discovering suggests a fancy interaction between these elements in modulating the organic pathways of ELGs.
In abstract, gentle clustering and KEGG pathway evaluation revealed that SD, significantly when mixed with SiNPs publicity, reprograms the temporal structure of the ELG transcriptome. These alterations mirror a fancy interplay between environmental and behavioral stressors and have profound implications for the regulation of lacrimal gland operate and circadian homeostasis.
3.7 Alterations in core clock genes of ELGs promoted by sleep deprivation in SiNPs-treated mice
To research the results of SiNPs and SD on the molecular circadian equipment of the lacrimal glands, we analyzed the temporal expression profiles of 12 core clock genes (Nr1d1, Nr1d2, Clock, Per1, Per2, Per3, Arntl, Cry1, Cry2, Npas2, Rora, and Rorc) throughout the NC, SiNPs-treated, and SD + SiNPs-treated teams(Fig. 8A). A two-way repeated measures ANOVA was carried out to judge the results of group, time level, and their interplay on gene expression ranges. The outcomes are summarized in Desk S15. A major major impact of time was noticed for Per3, Cry2, and Npas2, whereas no important impact of group or group × time interplay was discovered. These outcomes point out that gene expression different over time, per circadian regulation, however was not considerably affected by SiNPs or SD remedies. For Nr1d1, Arntl, Per1, Cry1, and Rora, important major results of time and group × time interplay had been detected, however not for group alone, indicating that temporal expression dynamics differed between remedies regardless of related total expression ranges. Though important major results of group, time, and group × time interplay had been noticed for Nr1d2, Clock, Per2, and Rorc, submit hoc comparisons between teams didn’t reveal statistically important variations. These findings recommend that therapy influenced the temporal expression dynamics of those genes, however variations between teams had been comparatively modest and didn’t mirror persistent modifications in common expression ranges. A separate two-way ANOVA confirmed that therapy, time level, and their interplay considerably influenced gene expression ranges. Unbiased-sample t-tests at particular person time factors confirmed that, though not all comparisons reached significance, each SiNPs and SD + SiNPs remedies independently or synergistically affected gene expression at a number of time factors. Moreover, the SD + SiNPs-treated group exhibited distinct expression patterns in comparison with the SiNPs-treated group at a number of time factors.
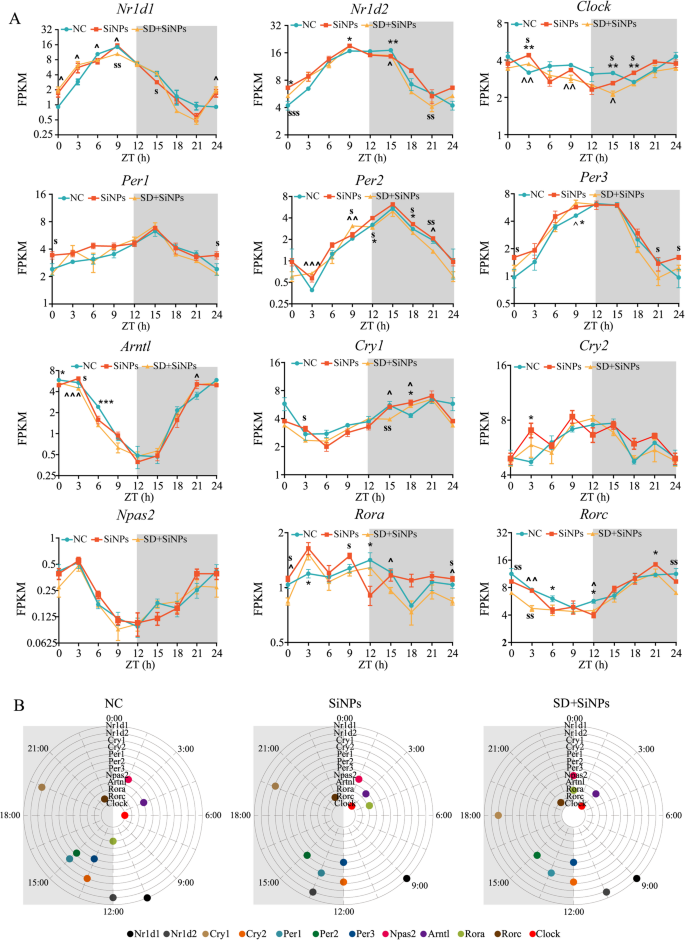
Influence of SiNPs and SD + SiNPs therapy on circadian transcription in murine ELGs. (A) The 24-hour expression profiles of 12 core clock genes, together with Nr1d1 (REV-ERBα), Nr1d2 (REV-ERBβ), Clock, Per1, Per2, Per3, Arntl (Bmal1), Cry1, Cry2, Npas2, Rora, and Rorc, are introduced. The x-axis represents the sampling time factors, whereas the y-axis signifies gene expression ranges at particular ZT factors. The inexperienced, orange, and yellow strains correspond to the NC, SiNPs-treated, and SD + SiNPs-treated teams, respectively. Grey shading denotes the darkish part of the LD cycle. Three animals per group had been sampled each three hours. At every time level, unbiased samples t-tests had been utilized to evaluate variations between the NC and SiNPs-treated teams, the NC and SD + SiNPs-treated teams, and the SiNPs-treated and SD + SiNPs-treated teams. Statistical significance is indicated as follows: *P < 0.05, **P < 0.01, ***P < 0.001 for comparisons between the NC and SiNPs-treated teams; ^P < 0.05, ^^P < 0.01, ^^^P < 0.001 for comparisons between the NC and SD + SiNPs-treated teams; and sP < 0.05, ssP < 0.01, sssP < 0.001 for comparisons between the SiNPs-treated and SD + SiNPs-treated teams. (B) The distribution of peak phases for core clock genes is proven for the NC, SiNPs-treated, and SD + SiNPs-treated teams. Grey shading represents the darkish part of the circadian cycle
Determine 8B illustrates the expression phases of the core clock genes throughout the three teams. Following SiNPs therapy and SD + SiNPs therapy, the expression phases of 11 out of 12 core clock genes, excluding Per2, had been altered relative to the NC group. Particularly, the phases of Nr1d1, Nr1d2, Clock, Per1, Per3, and Cry2 had been affected by SiNPs therapy and remained per these of the SiNPs-treated group following SD + SiNPs therapy. The phases of Rorc, Arntl, Cry1, and Npas2 remained unchanged after SiNPs therapy however shifted following SD + SiNPs therapy. The part of Rora was influenced by each remedies. Particularly, within the NC group, 4 clock parts are current within the gentle cycle, exhibiting 4 peak phases: 01:30 (Npas2), 04:30 (Artnl), 06:00 (Clock), and 10:30 (Nr1d1). Six clock parts are discovered in the dead of night cycle, with peak phases at 13:30 (Cry2 and Per3), 15:00 (Per1 and Per2), 19:30 (Cry1), and 22:30 (Rorc). Moreover, two peak phases at 12:00 (Rora and Nr1d2). Moreover, within the SiNPs-treated group, 5 clock parts are distributed within the gentle cycle, with 4 peak phases: 01:30 (Npsa2), 03:00 (Clock and Artnl), 04:30 (Rora), and 09:00 (Nr1d1). At midnight cycle, 5 clock parts are current, exhibiting 4 peak phases at 13:30 (Per1 and Nr1d2), 15:00 (Per2), 19:30 (Cry1), and 22:30 (Rorc). On the transition between the sunshine and darkish cycles, two clock parts are noticed, peaking at 12:00 (Per3 and Cry2). Furthermore, within the SD + SiNPs-treated group, 4 clock parts are discovered within the gentle cycle, with three peak phases at 03:00 (Clock and Artnl), 09:00 (Nr1d1), and 10:30 (Nr1d2). At midnight cycle, 4 clock parts present peak phases at 13:30 (Per1), 15:00 (Per2), 18:00 (Cry1), and 22:30 (Rorc). On the junction of the sunshine and darkish cycles, 4 clock parts peak at 00:00 (Npas2 and Rora) and 12:00 (Cry2 and Per3).
Collectively, these findings recommend that whereas SiNPs and SD + SiNPs remedies don’t drastically alter the common expression ranges of core circadian genes, they considerably have an effect on their part relationships and temporal dynamics. This reprogramming of clock gene oscillations highlights a nuanced but essential mode of circadian disruption inside the ELGs beneath environmental and behavioral stress.
3.8 Immune cell infiltration triggered by sleep deprivation in SiNPs-treated mice
Leukocyte migration into peripheral tissues is tightly regulated by circadian rhythms [49]. To look at the influence of SD on the immune microenvironment of the ELGs, the temporal dynamics of CD4⁺ and CD8⁺ T cell populations had been analyzed over a 24-hour interval. A major enhance in CD4⁺ T cell numbers was noticed between the SD + SiNPs-treated group and the NC group, with SD altering the height time of CD4⁺ T cell accumulation within the ELGs (Figs. 9A, C-D). Related tendencies had been noticed for CD8⁺ T cells (Figs. 9B, E-F).
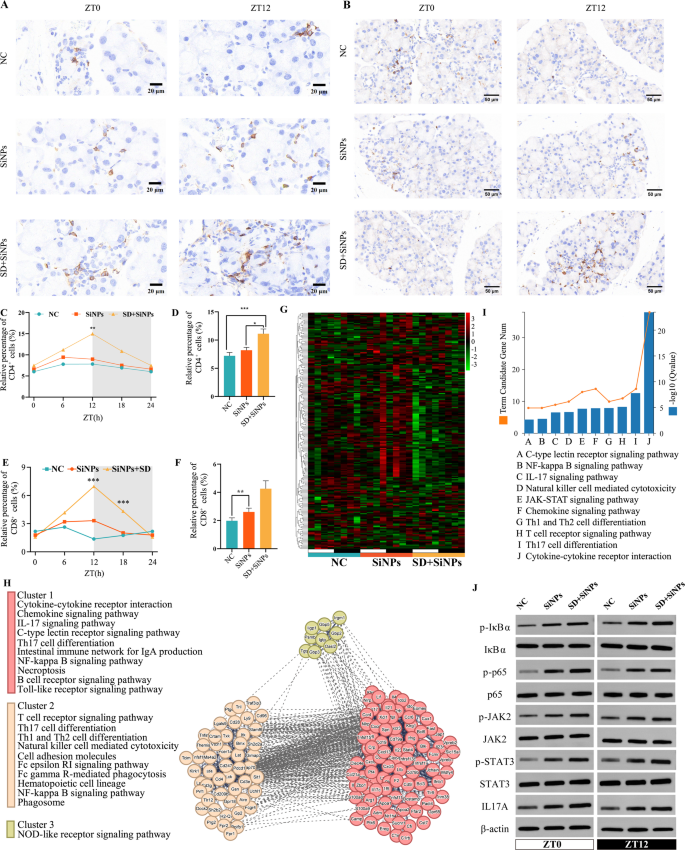
Influence of SiNPs and SD + SiNPs therapy on immune cells and genes in murine ELGs. (A) Consultant immunohistochemical photographs of CD4+ T cells in murine ELGs at ZT0 and ZT12, from NC, SiNPs-treated and SiNPs + SD-treated teams. Scale bar: 20 μm. (B) Consultant immunohistochemical photographs of CD8+ T cells in murine ELGs at ZT0 and ZT12, from the NC, SiNPs-treated, and SD + SiNPs-treated teams. Scale bar: 50 μm. (C) Quantitative evaluation of CD4+ T cell in murine ELGs, evaluating the diurnal variation of optimistic cell ratio among the many NC group, the SiNPs-treated group and the SD + SiNPs-treated group. **P < 0.01. (D) Common abundance of CD4+ T cell in murine ELGs from the NC, the SiNPs-treated and the SD + SiNPs-treated teams. Statistical evaluation was carried out utilizing the Kruskal–Wallis take a look at (non-parametric), adopted by Dunn’s submit hoc take a look at for a number of comparisons. *P < 0.05, ***P < 0.001. (E) Quantitative evaluation of CD8+ T cell in murine ELGs, evaluating the diurnal variation of optimistic cell ratio amongst NC group, SiNPs-treated group and SD + SiNPs-treated group. For NC group, P = 0.3391. For SiNPs-treated group, P = 0.4931. For SD + SiNPs-treated group, P = 0.0001. *P < 0.05, **P < 0.01, ***P < 0.001. NS: not important. (F) Common abundance of CD8+ T cells in murine ELGs from NC, SiNPs-treated and SD + SiNPs-treated teams. **P < 0.01. (G) Heatmaps of diurnal expression for immune-related DEGs between the NC group and SiNPs-treated group in murine ELGs. The expression ranges of immune-related genes had been obtained from RNA-Seq and expression vary of DEGs was normalized to ± 3. (H) The PPINs and purposeful clusters (cluster 1–3) with related KEGG pathways of immune-related genes between the SiNPs-treated group and SD + SiNPs-treated group. (I) The highest 10 KEGG pathways enriched histogram of immune-related genes with P < 0.05 had been displayed. (J) Immunoblotting of phosphorylation of STAT3, JAK2, phosphorylation of IκBα and p65, and IL17A in ELGs at ZT0 and ZT12, from NC, SiNPs-treated and SD + SiNPs-treated teams
Additional evaluation of immune-related gene expression revealed important transcriptional alterations within the SiNPs-treated group in comparison with the NC group, with SD additional exacerbating these modifications, as illustrated by the heatmap (Fig. 9G). In comparison with NC group, SiNPs-treated group exhibited 134 upregulated and 69 downregulated genes, whereas SD + SiNPs-treated group confirmed 81 upregulated and 122 downregulated genes (Desk S16). Protein-protein interplay community (PPIN) evaluation highlighted the central roles of a number of core genes inside these pathways (Fig. 9H), suggesting their potential involvement as key regulatory elements in SD- and particulate matter (PM)-induced lacrimal gland injury. Moreover, KEGG pathway enrichment evaluation recognized key immune pathways, together with the NF-κB signaling pathway, IL-17 signaling pathway, JAK-STAT signaling pathway, chemokine signaling pathway, and Th1/Th2 cell differentiation (Fig. 9I and Desk S17). To additional consider the activation standing of those pathways, we measured particular molecular markers: phosphorylation of STAT3 and JAK2 for the JAK-STAT pathway, phosphorylation of IκBα and p65 for the NF-κB pathway, and IL-17A expression ranges for the IL-17 signaling pathway. As proven in Fig. 9J and Figures S3-S4, SiNPs considerably elevated the phosphorylation of STAT3, JAK2, IκBα, and p65, in addition to the expression of IL-17A. Moreover, SD augmented these SiNPs-induced will increase (Fig. 9J and Figures S3–S4). The dysregulation of those key immune pathways seemingly contributes to power irritation and disruption of immune homeostasis, thereby exacerbating lacrimal gland damage and dysfunction.
Collectively, these outcomes display that SD exacerbates SiNPs-induced immune perturbations by enhancing T cell infiltration and amplifying pro-inflammatory signaling within the lacrimal gland. The mixed activation of JAK-STAT, NF-κB, and IL-17A pathways suggests a mechanism for sustained immune activation and tissue damage, contributing to the development of dry eye pathology.
3.9 Neuro-related transcriptome profile alterations within the ELGs triggered by sleep deprivation in SiNPs-treated mice
To judge the influence of SD and SiNPs publicity on neural features in murine ELGs, nerve-related genes had been recognized and analyzed. A complete of 98 upregulated and 74 downregulated DEGs had been recognized between the NC and SiNPs-treated teams, and 96 upregulated and 75 downregulated DEGs between the NC and SD + SiNPs-treated teams (Desk S18). The heatmap illustrates distinct expression patterns of neuro-related genes among the many NC, SiNPs-treated, and SD + SiNPs-treated teams inside the LD cycle (Fig. 10A). KEGG enrichment evaluation recognized the highest 10 considerably enriched pathways, primarily related to sign transduction, circadian rhythm synchronization, and neural regulation (Fig. 10C and Desk S19).
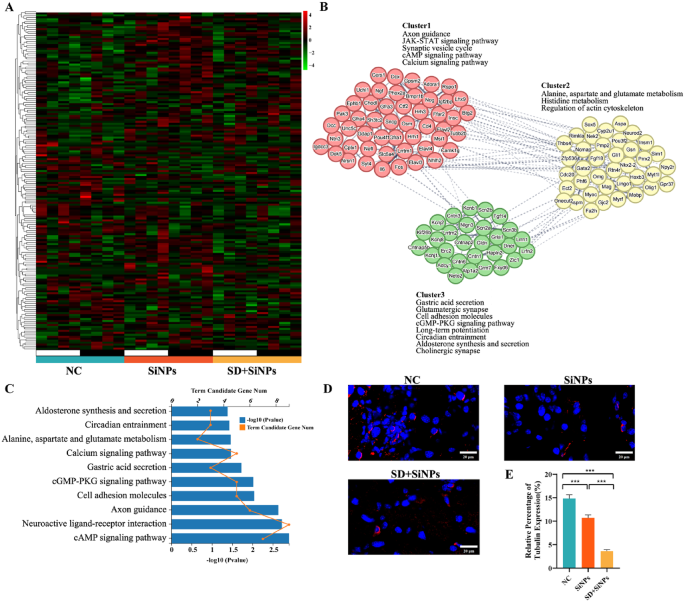
Immune alterations in murine ELGs following SiNPs and SD + SiNPs remedies. (A) Heatmaps of diurnal expression for nerve-related DEGs between the NC group and SiNPs-treated group in murine ELGs. The expression ranges of immune-related genes had been obtained from RNA-Seq and expression vary of DEGs was normalized to ± 4. (B) The PPINs and purposeful clusters (cluster 1–3) with related KEGG pathways of nerve-related genes between the NC group and SiNPs-treated group. (C) The highest 10 KEGG pathways enriched histogram of nerve-related genes with P < 0.05 had been displayed. (D) Consultant photographs of anti-β-III tubulin immunostaining in ELGs from the NC, SiNPs, and SD + SiNPs teams. Scale bar:20 μm. (E) Quantitative evaluation of anti-β-III tubulin staining in ELGs from the NC, SiNPs, and SD + SiNPs teams. Statistical evaluation was carried out utilizing Brown-Forsythe ANOVA. A number of comparisons had been performed utilizing the Video games-Howell submit hoc take a look at. For NC vs. SiNPs, P = 0.0007. For NC vs. SD + SiNPs, P < 0.0001. For SiNPs vs. SD + SiNPs, P < 0.0001. ***P < 0.001
To additional discover the interactions amongst these genes, PPINs had been constructed, categorizing neuro-related genes into three clusters (Fig. 10B). In line with KEGG purposeful annotation, SiNPs publicity could induce neurological dysfunction in ELGs, characterised by disruptions in sign transduction pathways, inhibition of neurogenesis, and neurotransmitter cycle issues.
Immunofluorescence evaluation of β-III tubulin, a neuronal marker, revealed decreased sign depth within the ELGs of the SiNPs-treated and SD + SiNPs-treated mice, indicating decreased innervation or neurodegeneration (Fig. 10D). Quantification confirmed a big discount within the β-III tubulin-positive density within the SiNPs-treated group in comparison with the NC group, and was additional diminished beneath SD (Fig. 10E). These findings recommend that SiNPs compromise the neural density of the ELGs in mice, doubtlessly contributing to decreased tear secretion. SD intensifies SiNPs-induced neural injury, thereby selling lacrimal dysfunction and dry eye development.
3.10 ROS accumulation and DNA injury within the ELGs triggered by sleep deprivation in SiNPs-treated mice
To look at the influence of SD on oxidative stress within the ELGs, immunofluorescence staining for ROS was carried out. In comparison with the NC group, the SiNPs-treated group exhibited a big enhance in ROS accumulation (P < 0.01). This enhance was additional amplified by SD, as evidenced by enhanced fluorescence depth (Figs. 11A-C). We additionally measured MDA ranges, a marker of lipid peroxidation, within the ELGs. In step with the ROS findings, MDA ranges had been considerably elevated within the SiNPs group in comparison with the NC group (P < 0.001). SD additional augmented this enhance in MDA (Figs. 11D-E). Moreover, each SiNPs publicity and the mixed SiNPs + SD therapy considerably disrupted oscillatory rhythms in contrast with the NC group (Figs. 11C and E).
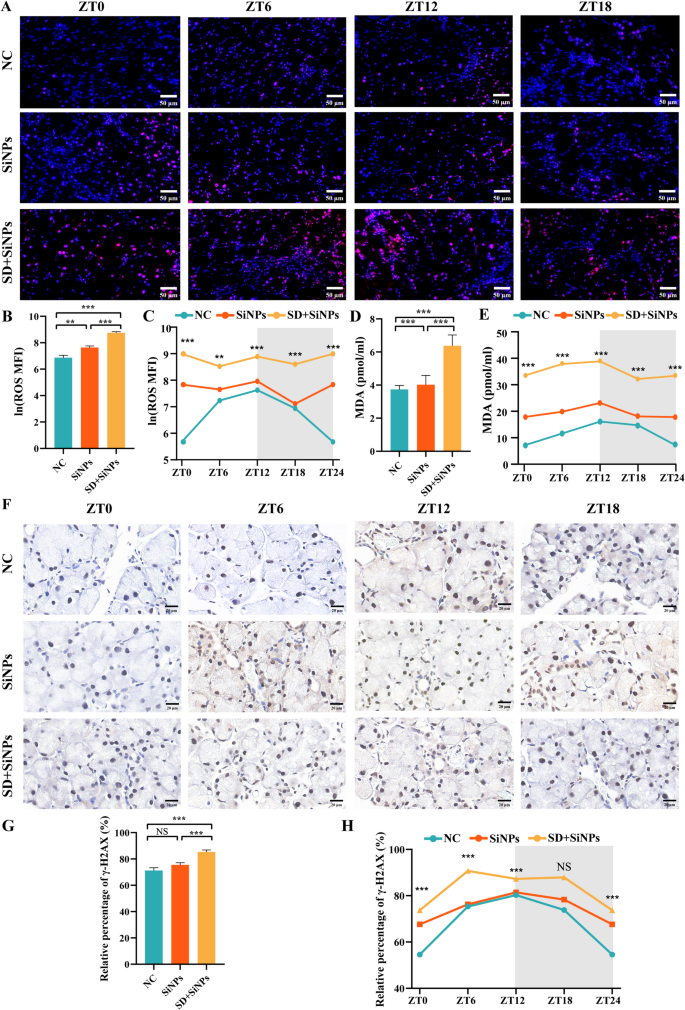
Oxidative stress and DNA injury in murine ELGs following SiNPs and SD + SiNPs remedies. (A) Consultant photographs exhibiting ROS ranges at ZT0, ZT6, ZT12, and ZT18 in murine ELGs from the NC, SiNPs-treated, and SD + SiNPs-treated teams. Scale bar: 50 μm. (B) Quantitative evaluation of ROS ranges in murine ELGs from NC, SiNPs-treated, and SD + SiNPs-treated teams. Imply fluorescence depth (MFI) values of ROS had been log-transformed utilizing the pure logarithm (ln) to enhance normality earlier than statistical evaluation. Statistical evaluation was carried out utilizing Brown-Forsythe ANOVA. A number of comparisons had been performed utilizing the Video games-Howell submit hoc take a look at. For NC vs. SiNPs, P = 0.002. For NC vs. SD + SiNPs, P < 0.0001. For SiNPs vs. SD + SiNPs, P < 0.0001. **P < 0.01, ***P < 0.001. (C) Diurnal modifications of ROS ranges in murine ELGs from NC, SiNPs-treated, and SD + SiNPs-treated teams. Statistical analyses had been carried out on pure log-transformed knowledge. For the NC group, F = 19.65, P < 0.0001. For the SiNPs-treated group, F = 3.794, P = 0.0265. For the SD + SiNPs-treated group, F = 1.957, P = 0.1499. **P < 0.01, ***P < 0.001. (D) Quantitative evaluation of MDA ranges in murine ELGs from the NC, SiNPs-treated, and SD + SiNPs-treated teams. Statistical evaluation was carried out utilizing Brown-Forsythe ANOVA. A number of comparisons had been performed utilizing the Video games-Howell submit hoc take a look at. For NC vs. SiNPs, P < 0.001. For NC vs. SD + SiNPs, P < 0.001. For SiNPs vs. SD + SiNPs, P < 0.001. ***P < 0.001. (E) Diurnal modifications of MDA ranges in murine ELGs from the NC, SiNPs, SD + SiNPs. Statistical analyses had been carried out on pure log-transformed knowledge. For the NC group, F = 57.601, P < 0.001. For the SiNPs-treated group, F = 4.732, P = 0.012. For the SD + SiNPs-treated group, F = 2.559, P = 0.084. ***P < 0.001. (F) Consultant photographs exhibiting γ-H2AX ranges at ZT0, ZT6, ZT12, and ZT18 in murine ELGs from the NC, SiNPs-treated, and SD + SiNPs-treated teams. Scale bar: 20 μm. (G) Quantitative evaluation of γ-H2AX ranges in murine ELGs from the NC, SiNPs-treated, and SD + SiNPs-treated teams. Statistical evaluation was carried out utilizing the Kruskal–Wallis take a look at (non-parametric), adopted by Dunn’s submit hoc take a look at for a number of comparisons. For NC vs. SiNPs, P = 0.8374. For NC vs. SD + SiNPs, P < 0.0001. For SiNPs vs. SD + SiNPs, P = 0.0006. ***P < 0.001. NS: not important. (H) Diurnal modifications of γ-H2AX ranges in murine ELGs from the NC, SiNPs-treated, and SD + SiNPs-treated teams. For the NC group, F = 32.35, P < 0.0001. For the SiNPs-treated group, F = 7.167, P = 0.0017. For the SD + SiNPs-treated group, F = 20.02, P < 0.0001. ***P < 0.001. NS, not important
To additional consider the influence of oxidative stress on DNA injury, immunohistochemical staining for γ-H2AX, a marker of DNA double-strand breaks, was performed. The SD + SiNPs-treated group exhibited considerably larger γ-H2AX-positive staining in comparison with the NC group, indicating that ROS accumulation contributed to DNA injury within the ELGs (Figs. 11F-G). Furthermore, the oscillatory rhythm of γ-H2AX ranges was disrupted by SiNPs therapy and completely modified by SD + SiNPs therapy (Fig. 11H).
Taken collectively, these findings display that SiNPs publicity induces oxidative stress within the lacrimal glands, characterised by elevated ROS accumulation, lipid peroxidation, and DNA injury. Importantly, SD considerably amplifies this oxidative damage and disrupts circadian redox homeostasis, seemingly contributing to the structural and purposeful decline of ELGs.
3.11 Activation of NLRP3 inflammasome triggered by sleep deprivation in SiNPs-treated mice
The NLRP3 inflammasome performs a essential position in irritation and oxidative stress. To research the involvement of the ROS/NLRP3 pathway in SiNPs-induced dry eye exacerbated by SD, we evaluated the expression patterns of NLRP3 and its adaptor protein ASC in ELGs. Immunohistochemical staining of NLRP3+ cells was illustrated in Fig. 12A. Evaluation of NLRP3 expression revealed that SiNPs therapy alone didn’t considerably have an effect on NLRP3 manufacturing in ELGs, whereas a marked enhance was noticed when SiNPs had been mixed with SD (Fig. 12B), suggesting a attainable synergistic impact between SiNPs therapy and SD therapy. Determine 12C confirmed the oscillation of NLRP3+ cells within the NC, the SiNPs-treated, and the SD + SiNPs-treated teams. SiNPs therapy altered the every day oscillation of NLRP3 in comparison with the NC group, with each teams peaking at ZT12, however exhibiting totally different trough values (ZT6 within the NC group vs. ZT18 within the SiNPs group). SD therapy additional disrupted the every day rhythm of NLRP3, with the height occurring at ZT6 and the trough at ZT18. Moreover, ASC ranges had been considerably elevated within the SiNPs-treated group in comparison with the NC group (Figs. 12E-I), with its oscillatory rhythm maintained (Fig. 12D). SD additional elevated ASC ranges and disrupted its every day rhythmicity.
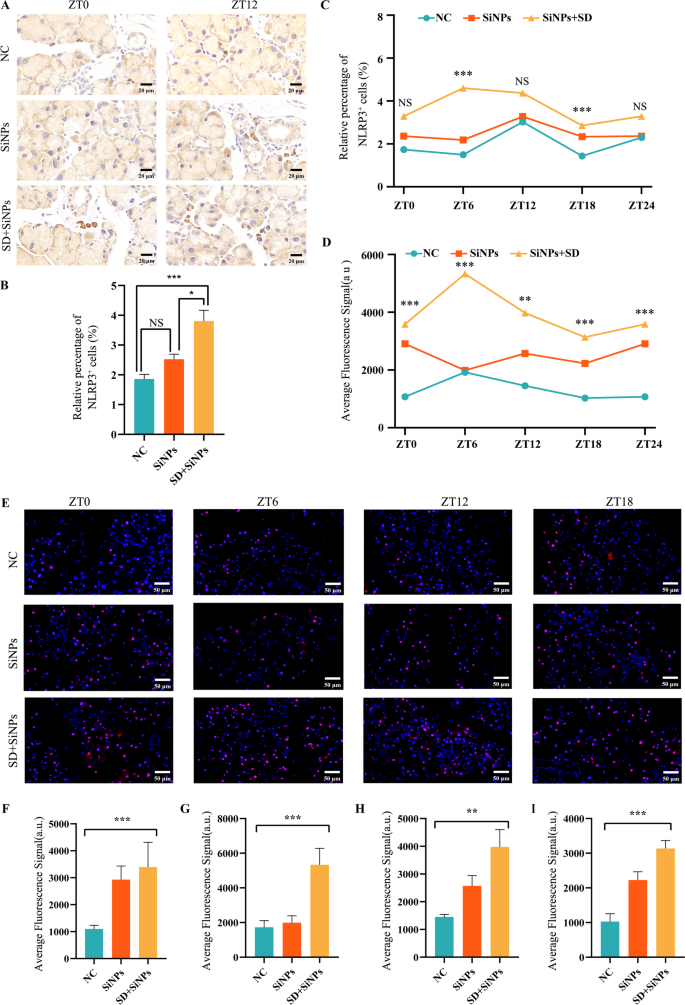
SD potentiates SiNPs-induced NLRP3 inflammasome activation in mouse ELGs. (A) Consultant immunohistochemical photographs of NLRP3+ cells in mouse ELGs at ZT0 and ZT12 for the NC, SiNPs-treated, and SD + SiNPs-treated teams. Scale bar: 20 µm. (B) Common abundance of NLRP3+ cells of ELGs from NC, SiNPs-treated, and SD + SiNPs-treated teams. Statistical evaluation was carried out utilizing the Kruskal–Wallis take a look at (non-parametric), adopted by Dunn’s submit hoc take a look at for a number of comparisons. Dunn’ s take a look at confirmed that the SD + SiNPs-treated group differed considerably from the NC group (***P < 0.001) and the SiNPs-treated group (*P < 0.05), whereas the NC group, and the SiNPs-treated group weren’t considerably totally different (NS). (C) Diurnal variation evaluation of NLRP3+ cell ratio in murine ELGs throughout the NC, SiNPs-treated group, and the SD + SiNPs-treated teams. Statistical evaluation confirmed no important diurnal variations in any group (ZT0: F = 1.419, P = 0.2665; ZT6: F = 19.05, P < 0.001; ZT12: F = 1.227, P = 0.3134; ZT18: F = 17.68, P < 0.001). ***P < 0.001. NS: not important. (D) Diurnal variation evaluation of ASC+ cells ratio in murine ELGs throughout NC group, SiNPs-treated group, and SD + SiNPs-treated group. For ZT0, F = 11.50, P = 0.0005; For ZT6, F = 10.98, P = 0.0008; For ZT12, F = 1.227, P = 0.005; For ZT18, F = 17.68, P < 0.0001. **P < 0.01, ***P < 0.001. (E) Consultant immunofluorescence photographs of ASC+ cells in mouse ELGs at ZT0, ZT6, ZT12, and ZT18 time factors for the NC group, the SiNPs-treated group, and the SD + SiNPs-treated group. Scale bar: 50 μm. (F-I) Quantitative evaluation of common fluorescence sign depth of ASC in mouse ELGs at ZT0 (G), ZT6 (H), ZT12 (I), and ZT18 (J) for the NC group, SiNPs-treated group, and SD + SiNPs-treated group. Statistical evaluation revealed important variations in any respect time factors: ZT0 (F = 11.50, P = 0.0005), ZT6 (F = 10.98, P = 0.0008), ZT12 (F = 1.227, P = 0.005), and ZT18 (F = 17.68, P < 0.0001). **P < 0.01, ***P < 0.001
Taken collectively, these findings recommend that SiNPs set off a light activation of the NLRP3 inflammasome, which SD amplifies whereas deranging the rhythmic expression of NLRP3 and ASC, presumably through ROS build-up and immune imbalance.

